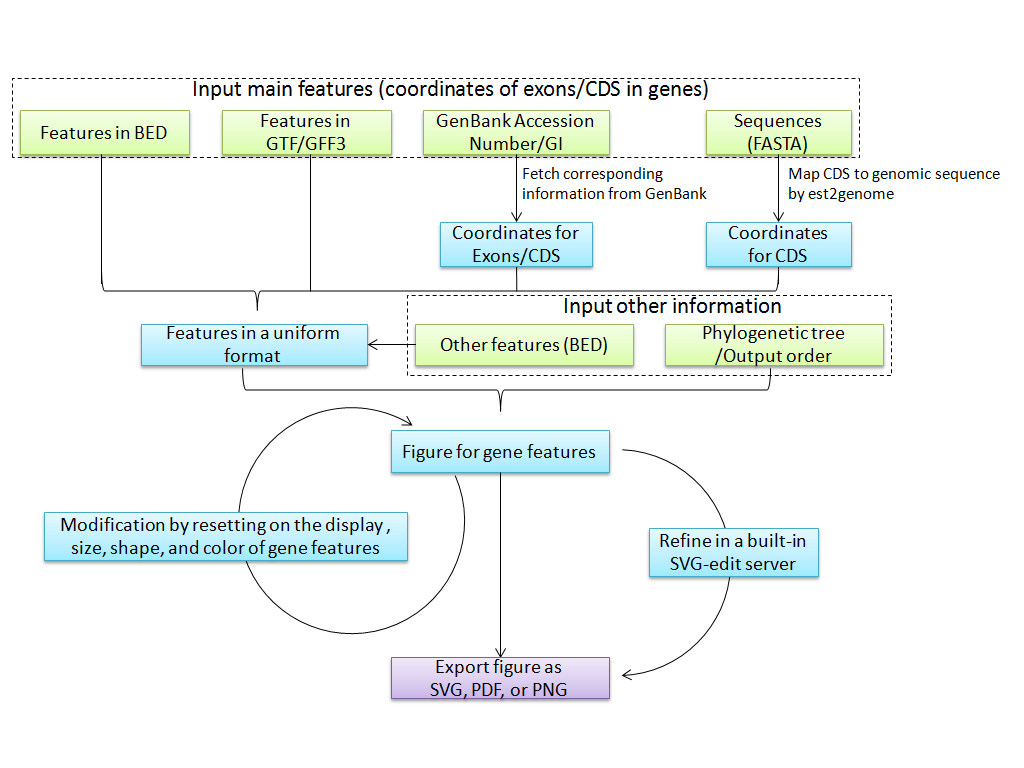
1. Work Flow of GSDS 2.0
GSDS 2.0 is designed for the visualization of annotated features for genes, and the generation of high-quality figures for publication. Besides the main features (i.e., coordinates of exons/CDS), other annotated features such as conserved elements and binding sites can also be displayed. GSDS 2.0 supports features in four types of formats, including BED, GTF/GFF3, GenBank Accession Number/GI, and Sequences in FASTA. After inputting these features, GSDS would transform them into a uniform format for figure generation. To facilitate evolutionary study, a phylogenetic tree can be uploaded and displayed on the side-panel of the figure.
 Download high-resolution work flow in PowerPoint
Download high-resolution work flow in PowerPoint
After the first generation of figures, users can turn on/off the display of specified features, and modify the sizes, shapes and colors of displayed features. Moreover, the generated figures can be sent to a built-in SVG-edit server for further refining. Finally, users can export the figure as SVG, PNG, or PDF.
2. Supported Gene Features and Formats
GSDS supports the display of main features (coordinates of exons/CDS), as well as any other features that can be described in BED formats, such as conserved elements, binding sites.
2.1 Input Main Features
Users can input the main features in four formats, including BED, GTF/GFF3, GenBank Accession Number/GI, and FASTA Sequences. Brief introduction for these formats are described as following:
2.1.1 BED Format
BED format uses the tab-separated fields to describe the annotated features (see UCSC for more details). Users can input features as following format:
geneID/transcriptID start end featureType phase(optional)
The first four columns are required (marked in blue), and the last column for intron phase is optional. To display the main feature correctly, the column for feature type should include exon, CDS or UTR.
Example:
AY077757 0 725 UTR . AY077757 725 1157 CDS 0 AY077757 1287 1757 CDS 2 AY077757 1867 1993 CDS 2 AY077757 2143 2303 CDS 0 AY077757 2303 2731 UTR .
Note: the coordinate system is zero-based in BED format.
2.1.2 GTF/GFF3 Format
The GTF/GFF3 is widely used to describe gene features, which contains 9 columns per line.
seqID source featureType start end score strand frame attributes(geneID and transcriptID required)
The columns in blue are required by GSDS 2.0, others are optional and can be '.', representing no value for this field.
Features other than exons,introns, UTRs,CDSs will also be displayed. If gene_name/transcript_name is provided, these will be shown in the graph instead of IDs.
Example for GTF (see more details)
chr . UTR 1 725 . + . gene_id "AY077757"; transcript_id "AY077757.1"; chr . CDS 726 1157 . + 0 gene_id "AY077757"; transcript_id "AY077757.1"; chr . CDS 1288 1757 . + 2 gene_id "AY077757"; transcript_id "AY077757.1"; chr . CDS 1868 1993 . + 2 gene_id "AY077757"; transcript_id "AY077757.1"; chr . CDS 2144 2303 . + 0 gene_id "AY077757"; transcript_id "AY077757.1"; chr . UTR 2304 2731 . + . gene_id "AY077757"; transcript_id "AY077757.1";
Example for GFF3 (see more details)
chr . gene 1 2731 . + . ID= AY077757 chr . mRNA 1 2731 . + . ID=AY077757.1;Parent=AY077757 chr . UTR 1 725 . + . ID=utr.1.AY077757;Parent=AY077757.1 chr . CDS 726 1157 . + 0 ID=cds.1.AY077757;Parent=AY077757.1 chr . CDS 1288 1757 . + 2 ID=cds.2.AY077757;Parent=AY077757.1 chr . CDS 1868 1993 . + 2 ID=cds.3.AY077757;Parent=AY077757.1 chr . CDS 2144 2303 . + 0 ID=cds.4.AY077757;Parent=AY077757.1 chr . UTR 2304 2731 . + . ID=utr.2.AY077757;Parent=AY077757.1
2.1.3 GenBank Accession Number/GI
Users can input GenBank accession number or GI. GSDS can fetch the corresponding information for exon/CDS coordinates from GenBank.

Example:
AY077757 AY514043 BK005038 BK005071 BK005073
2.1.4 FASTA Sequences
Users can input both the CDS sequences and genomic sequences in FASTA format. GSDS can map the CDS to genomic sequences by est2genome and get the coordinate of CDS.
Example for FASTA sequences:
>AY077757 ATGGGGGCGTTGGAGATATTAGATTACAACAACACTTTAGGAAAGAGAGACAGGGACTATGAAGTGAAGGAAGCGGCATGCATGGGAATACAAAACGCTA GGCAGCTGCTCCAGTCCCTGACGCAGGTGCGATCTCCAGTGGTGGACGAAGAATGCGATGTCATGGCTGGCGCTGCCATATCCAAGTTTCAGAAGGTGGT GTCACTACTGAGTCGCACTGGTCATGCACGGTTTCGTAGGAGAACGCGCAACGCTGCTGTTGCCGGTTACGCAGGCGTCTTCTTAGAGAGCTCCAACTTC TTCAGAGAAAATTCCCAGGAGACGTCGAGGGACAGAATCGTCTCGTCGGGCCATGCTAGCCCATCTCAGTTCACGCCGACGTCCTCGTCCAAGCCTCCTC AGTCACCTGAATTGCAGGCGATCAAATATAAGGTGTTTCCTCAAAGCTCTCGTTCCGCTGATGCGACGCCTGCCTCCAGTGACCCTGCTTCAGGAGTCCA TCATCCAAAGCCACTTCAGATCCTTCACAGCTCCATGATGCAGCAAAGCATTCCAGAACATATACTGCGTCCAGTGGCTAGTGCTGCGTATCGGCCAACT GCCCTTCCCCCGAATCCGTTCAACAAACAGGAGGTGGGCAGCAAGGAGGGGGTGAGCGGCCACAGTCCGGACAGTTCGTTGAGCTCAGGACCTCCGCAAT CAACTACAACGGCGTCGTTCCCAACCATGAGTGTGCAGGATGCGAGGATAACGAGCCTGCAGAATATGAAAACAGCCGAGCAACCTTCGGCGTTGCCCCC TCGCCCGCAGCCACCAACTCCCAAGAAAAAGTGCTCCGGGCAATCCGATGAGAACGGTGCAACTTGCGCAATCCTTGGCCGCTGCCATTGTTCAAAACGC AGGAAATTGCGGTTGAAGAGGACAATCACGGTTCGAGCAATCAGCAGCAAGTTGGCTGATATACCTTCGGATGAGTATTCATGGCGTAAGTATGGCCAGA AGCCTATCAAAGGATCACCACATCCGAGAGGATACTACAAGTGCAGCAGCATACGAGGCTGTCCAGCGAGAAAACACGTAGAGCGGTCAATGGAAGACTC ATCTATGTTGATTGTGACATACGAAGGCGATCATAACCATCCGCAATCGTCATCTGCTAATGGCGGATTAACAGTGCAGTCGCAATAG
Note: both the genomic sequences and CDS sequences are required.
2.2 Add Other Features
After inputting the main features, users can add other features in BED format as following:
geneID/transcriptID start end featureType
Example for other features:
AY077757 1928 1993 feature1 AY077757 2143 2261 feature1 AY514043 1062 1127 feature1 AY514043 1224 1342 feature2 BK005038 153 252 feature1 BK005038 1677 1851 feature1 BK005038 2464 2529 feature1 BK005038 2563 2729 feature2 BK005071 768 833 feature1 BK005071 927 1045 feature1 BK005073 1258 1435 feature1 BK005073 1777 1842 feature1 BK005073 1938 2050 feature2
3 Adding Phylogenetic Tree/Output Order
To facilitate evolutionary study, users can upload a phylogenetic tree for inputted genes in NEWICK format. GSDS will display it on the side-panel of gene features in the figure.
Example for phylogenetic tree:
(((AY514043,BK005071),BK005073),(AY077757,BK005038));
Note: A semicolon is required at the end of the tree. Branch lengths are ignored in GSDS.
Or choose the "output order" to adjust the output order of genes as following:
BK005038 BK005071 BK005073 AY077757 AY514043
if output order is provided, only the genes/transcripts included will be displayed.
4. Modifying the Generated Figure
Example for result page:
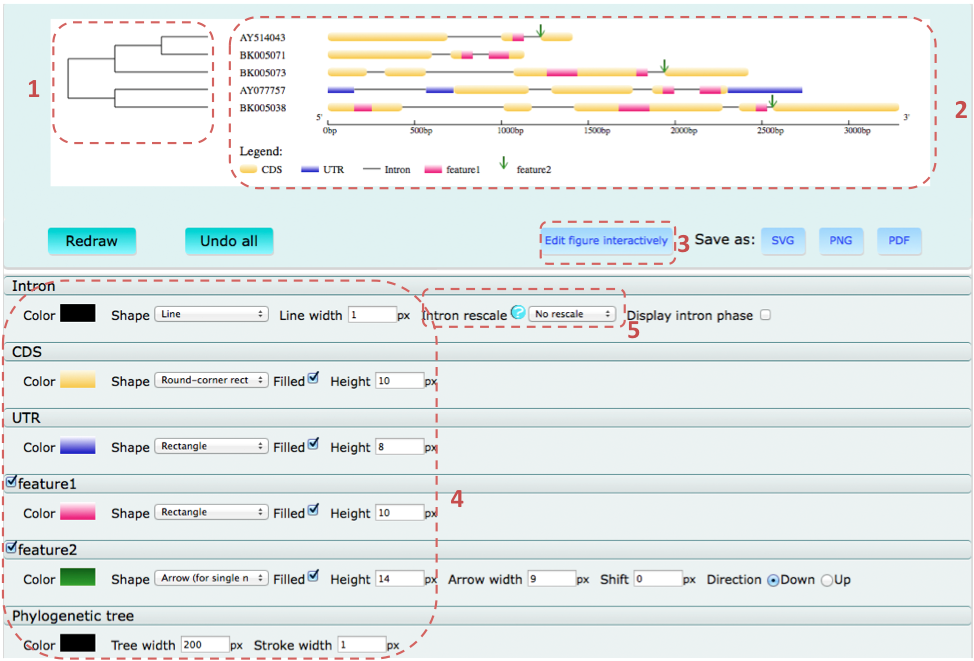
After the first generation of the figure, users can modify it by setting on the "region 4" in the result page, including turning off the display of specified features, modifying the color, shape, and size of the features, as well as rescaling the intron.
There are three options for "Intron rescale"
- No rescale: dispalying introns in the same scale with exons.
- Shrink: shrinking the length of the intron. In this case, a ratio should be specified, e.g., 0.01 means to shrink intron length to 1% of its original length.
- displaying all introns in the same length. In this case, features on intron would not be displayed.
Besides that, users can also send the generated figure to the built-in SVG-editor for further refining by clicking "Edit figure interactively".
Finally, users can save the figure as SVG, PNG, or PDF.
5. Installing a Local Version
Users can download the source code of GSDS 2.0 here, and install it locally(license). See manual for more information.




