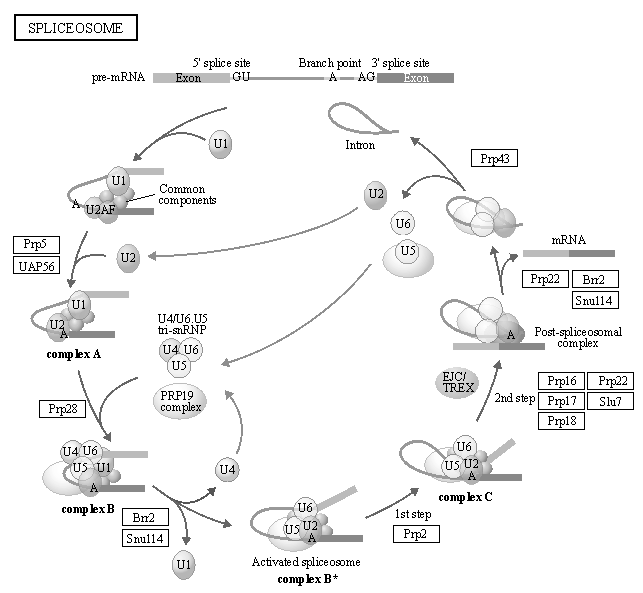
Pathway
KEGG PATHWAY is a collection of manually drawn pathway maps representing our knowledge of the molecular interaction, reaction and relation networks for:
1. Metabolism
2. Genetic Information Processing
3. Environmental Information Processing
4. Cellular Processes
5. Organismal Systems
6. Human Diseases
7. Drug Development.
Pathway annotation of 25 Fabaceae genomes were performed using GhostKoala, respectively, it is being a tool provided by the Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg).
Select a legume and click Submit to see the results of the gene annotation for the changed species. The relevant gene ids can be obtained from the Fabaceae blast and match under the Tools module.This link is Fabaceae blast and match.