Description
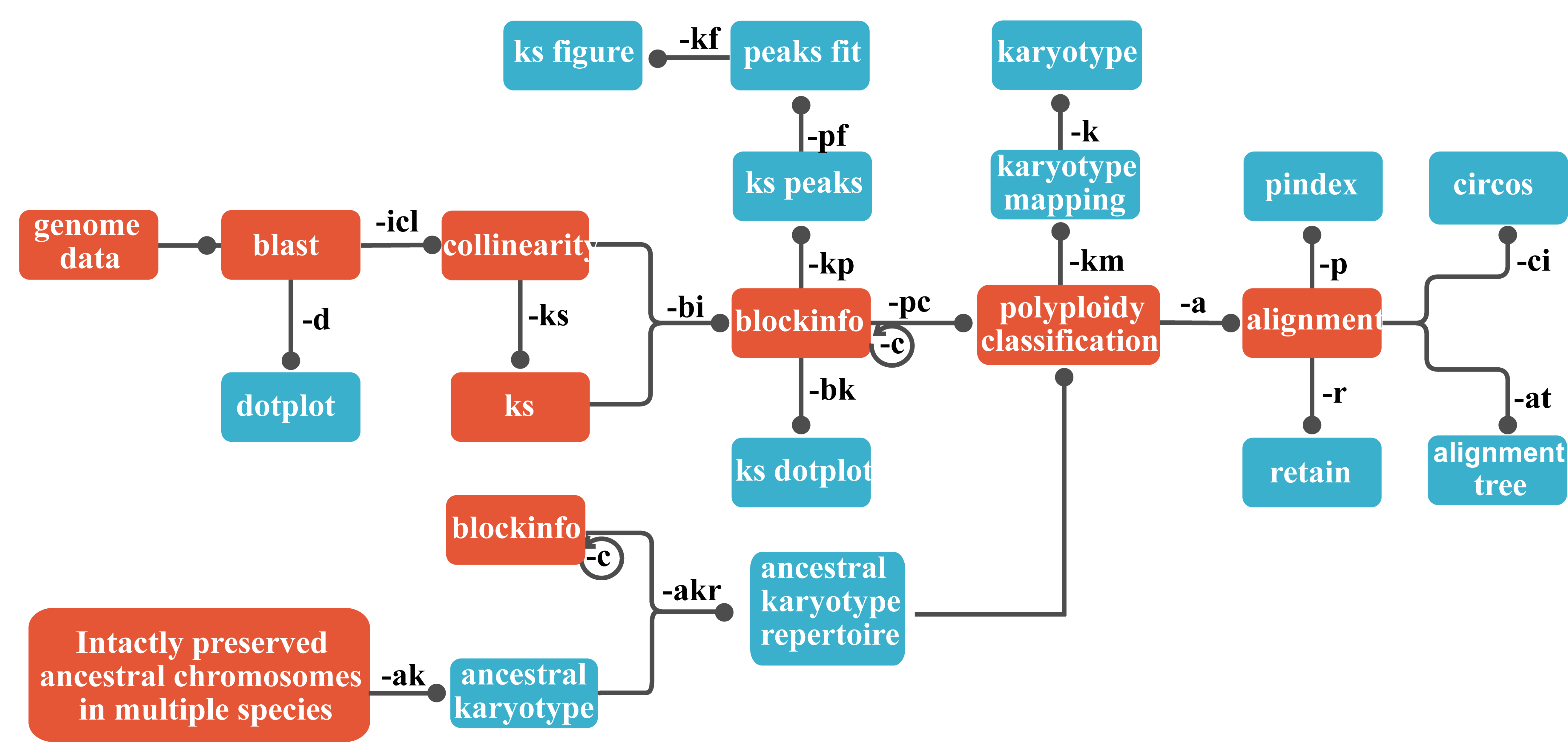
WGDI (Whole-Genome Duplication Integrated analysis), a Python-based command-line tool that facilitates comprehensive analysis of recursive polyploidization events and cross-species genome alignments. WGDI supports three main workflows (polyploid inference, hierarchical inference of genomic homology, and ancestral chromosome karyotyping) that can improve the detection of WGD and characterization of WGD-related events based on high-quality chromosome-level genomes. Significantly, it can extract complete synteny blocks and facilitate reconstruction of detailed karyotype evolution.
For detailed introductions to some usage functions, please refer to
https://wgdi.readthedocs.io/.
The latest and improved version download link is provided above.
Citating WGDI
Sun, P., Jiao, B., Yang, Y., Shan, L., Li, T., Li, X., … & Liu, J. (2022). WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Molecular plant, 15(12), 1841-1851.
https://doi.org/10.1016/j.molp.2022.10.018
Source Code Download
latest_wgdi: wgdi_protocol: