Dolplot
Dolplot Modular Function:Dolplot contains dot map and Ks distribution information between two species.
-
Dolplot
Select reference species and comparative species, and wait a few minutes to get the desired result.

Lists
Lists Modular Function:GGDB Lists contains colinear information of Gramineae species, which is divided into two modules: Colineartiy and Colineartiy D.
-
Colineartiy
Select reference species and comparative species, and wait a few minutes to get the desired result. Colineartiy module information is not involved in the cWGD incident.
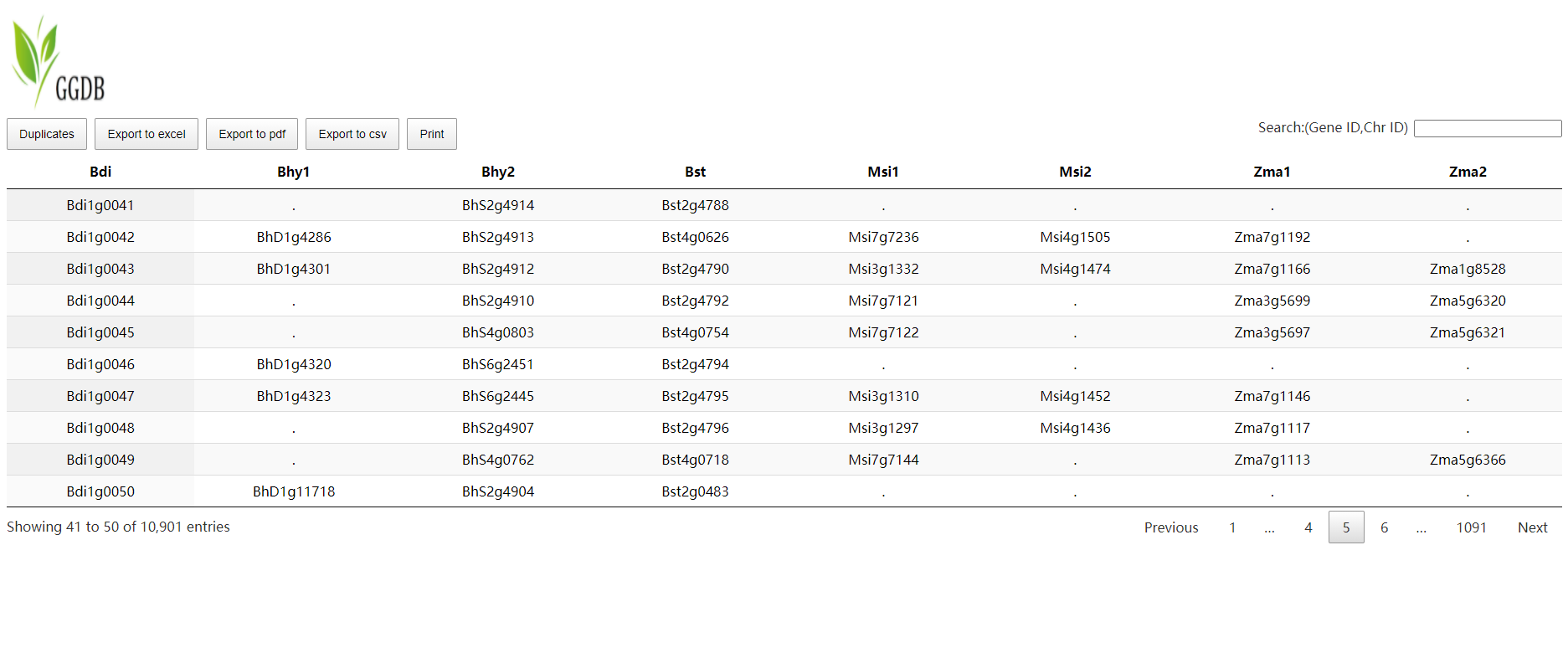
-
Colineartiy D
Select reference species and comparative species, and wait a few minutes to get the desired result. Colineartiy module information involves cWGD events.
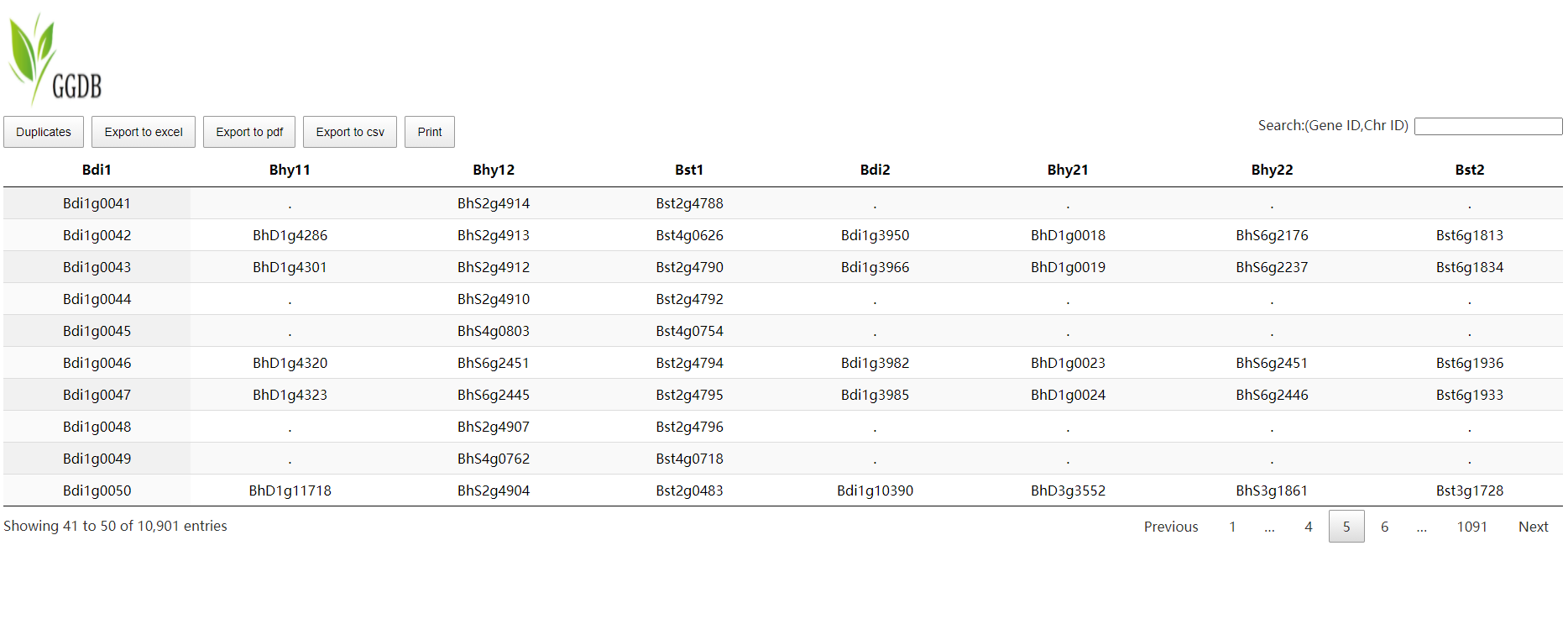
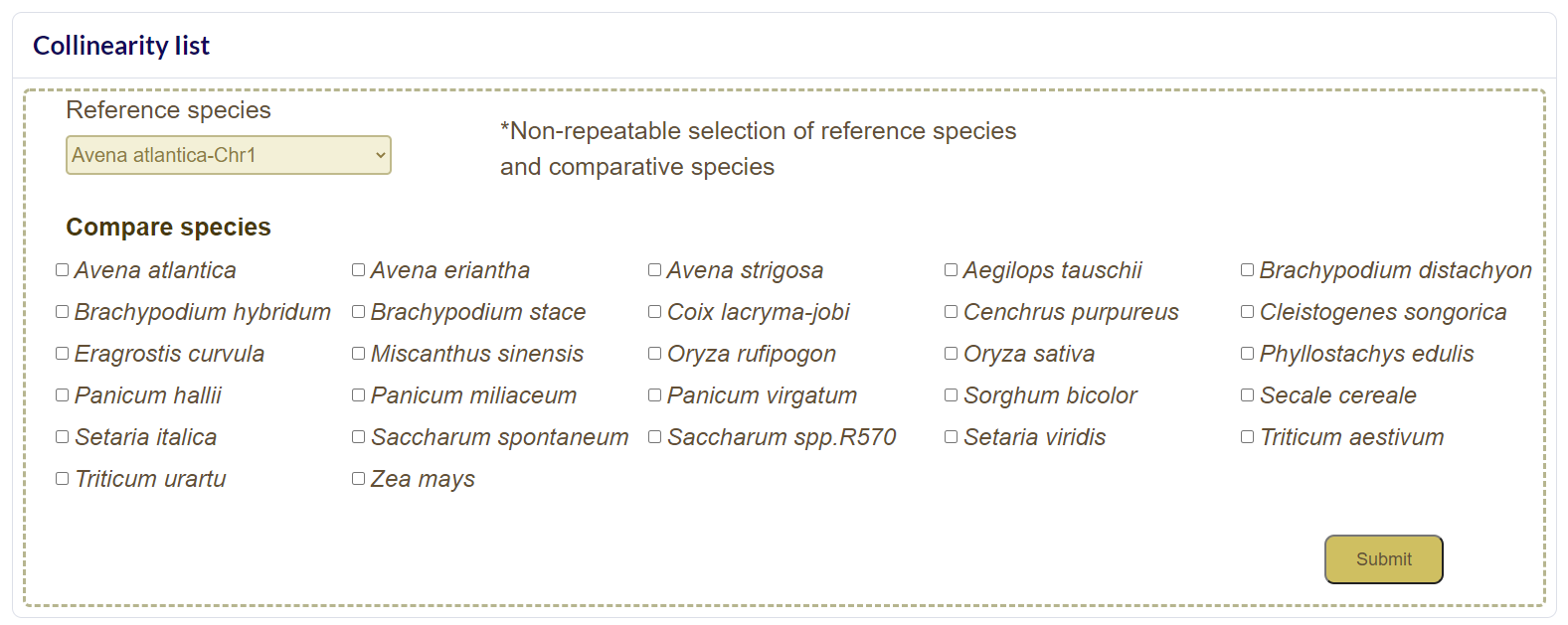
Pairwise Colineraity
Pairwise Colineraity Modular Function:Pairwise Colineraity can draw collinear maps between two species online, including two modules at chromosome level and gene level.
-
Chromosome Level
Select the reference species and the comparative species, and submit the chromosome number of the species to be compared, and wait a few minutes to get the desired result. Colineartiy is not involved in the cWGD incident.
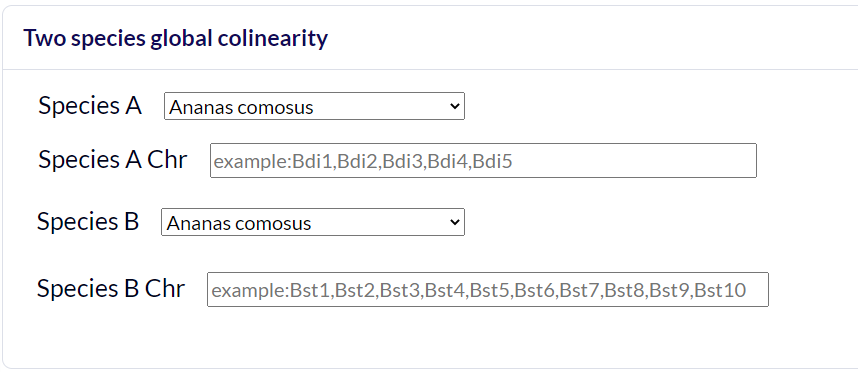
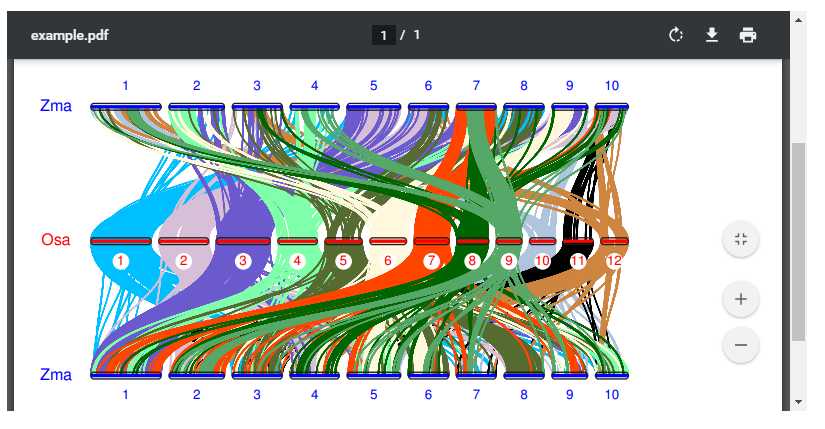
-
Gene Level
Select the reference species and compare species, and submit the genetic ID file to be mapped by the reference species, and wait a few minutes to get the desired result. Gene ID files can be obtained from the List module.

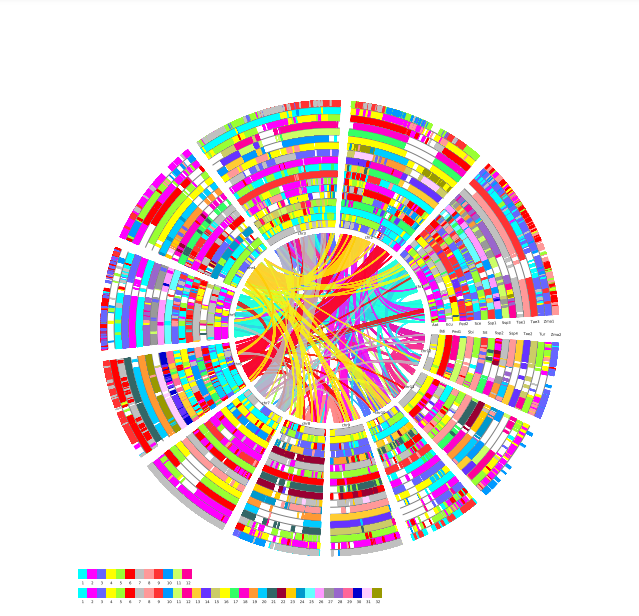
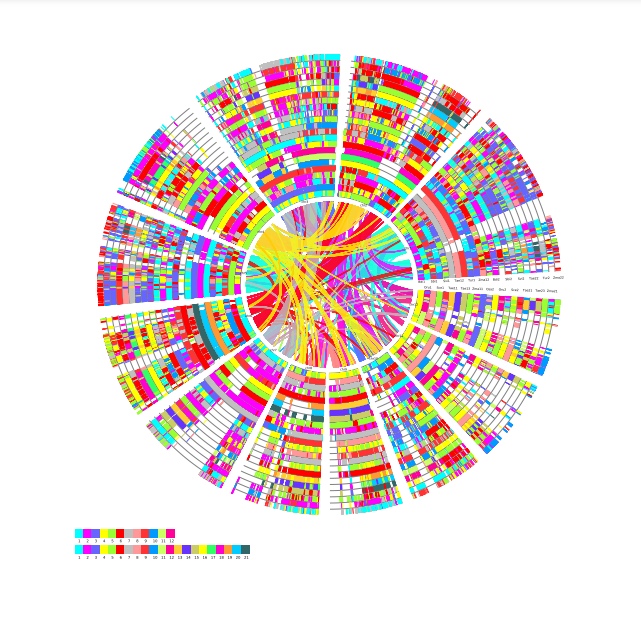
Local Colineraity
Local Colineraity Modular Function:The module can draw the local joint alignment map of multi-genomes, which is divided into two modules: Colinearity and Colineartiy D.
-
Colinearity
Select the reference species and compare species, submit the genetic ID file to be mapped by the reference species, and wait a few minutes to get the desired result. Colineartiy module information is not involved in the cWGD incident. Gene ID files can be obtained from the List module.
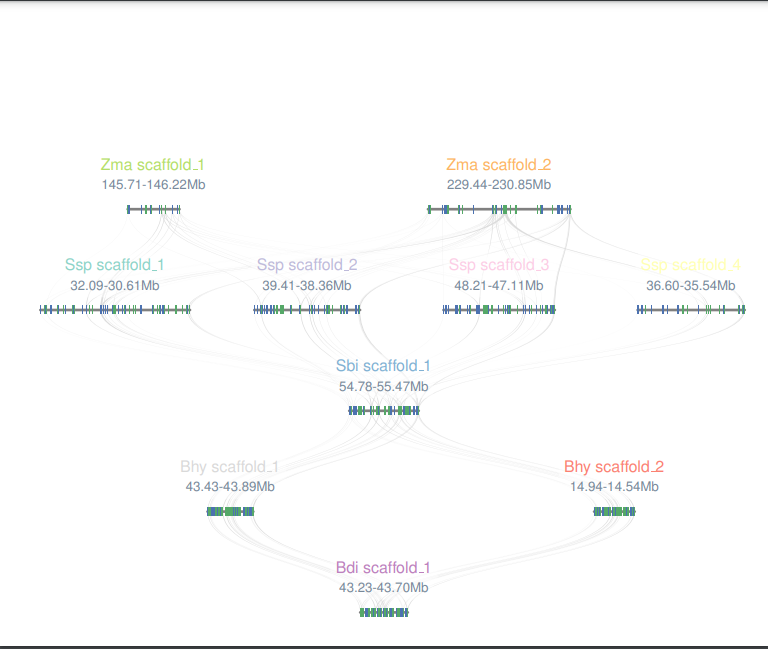
-
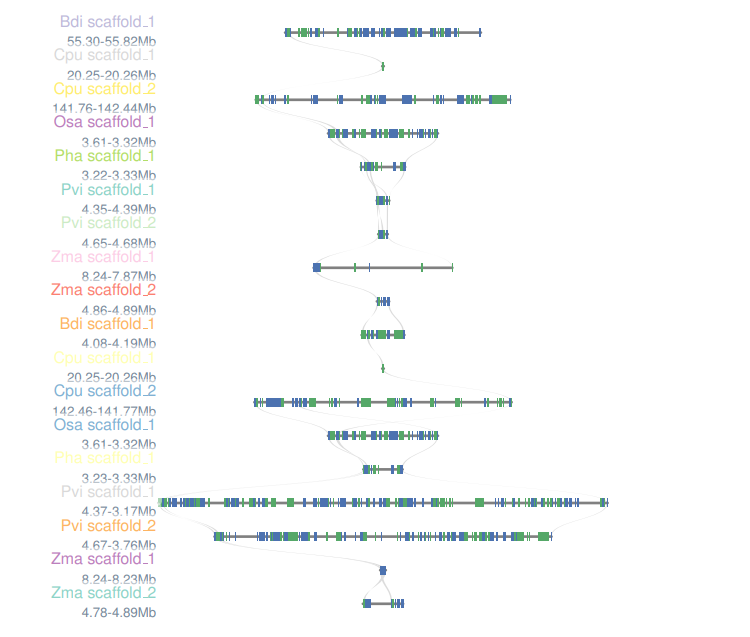
Colinearity D
Select the reference species and compare species, submit the genetic ID file to be mapped by the reference species, and wait a few minutes to get the desired result. Colineartiy module information is involved in the cWGD incident. Gene ID files can be obtained from the List D module.
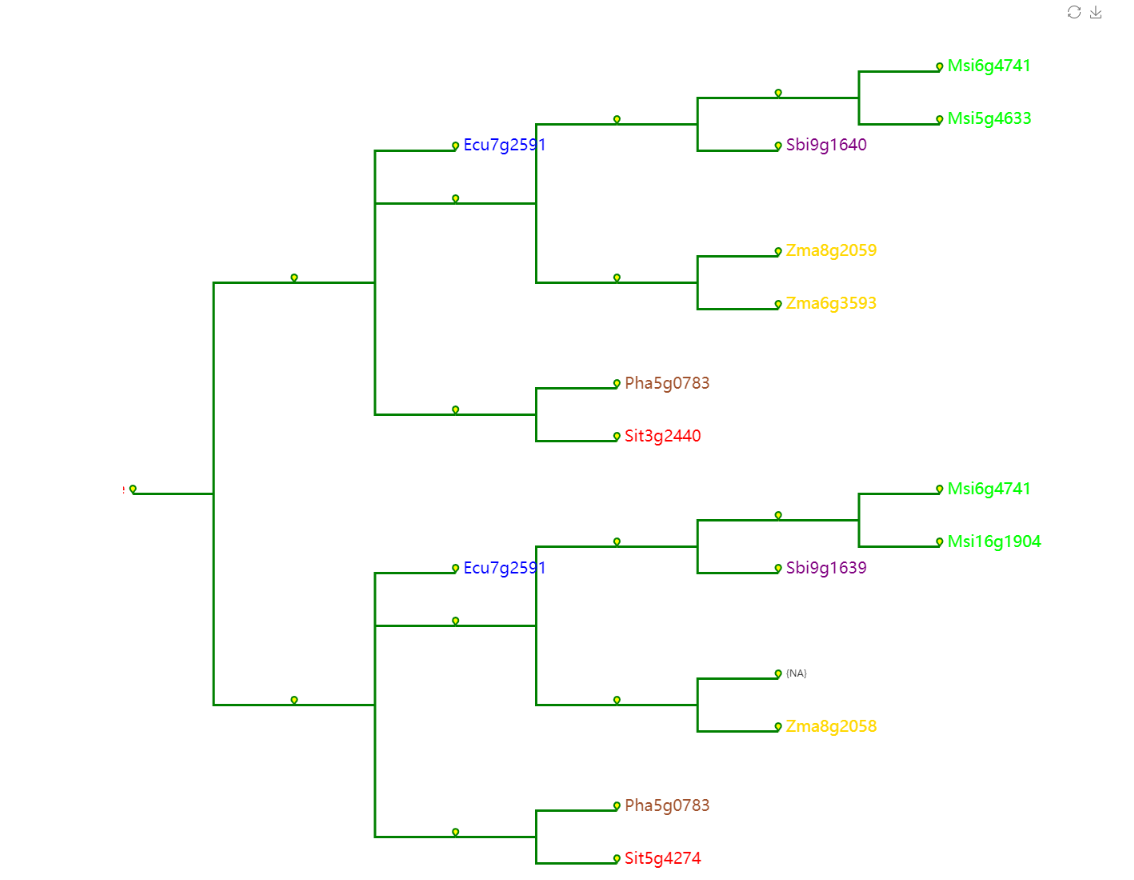
Trees
Trees Modular Function:Trees module can draw a single gene collinear evolution tree, including the real evolution rate evolution tree and the corrected gene evolution tree.
-
Actual Gene Tree
Select the reference species and compare species, submit the genetic ID file to be mapped by the reference species, and wait a few minutes to get the desired result. Colineartiy module information is involved in the cWGD incident. Gene ID files can be obtained from the List D module.
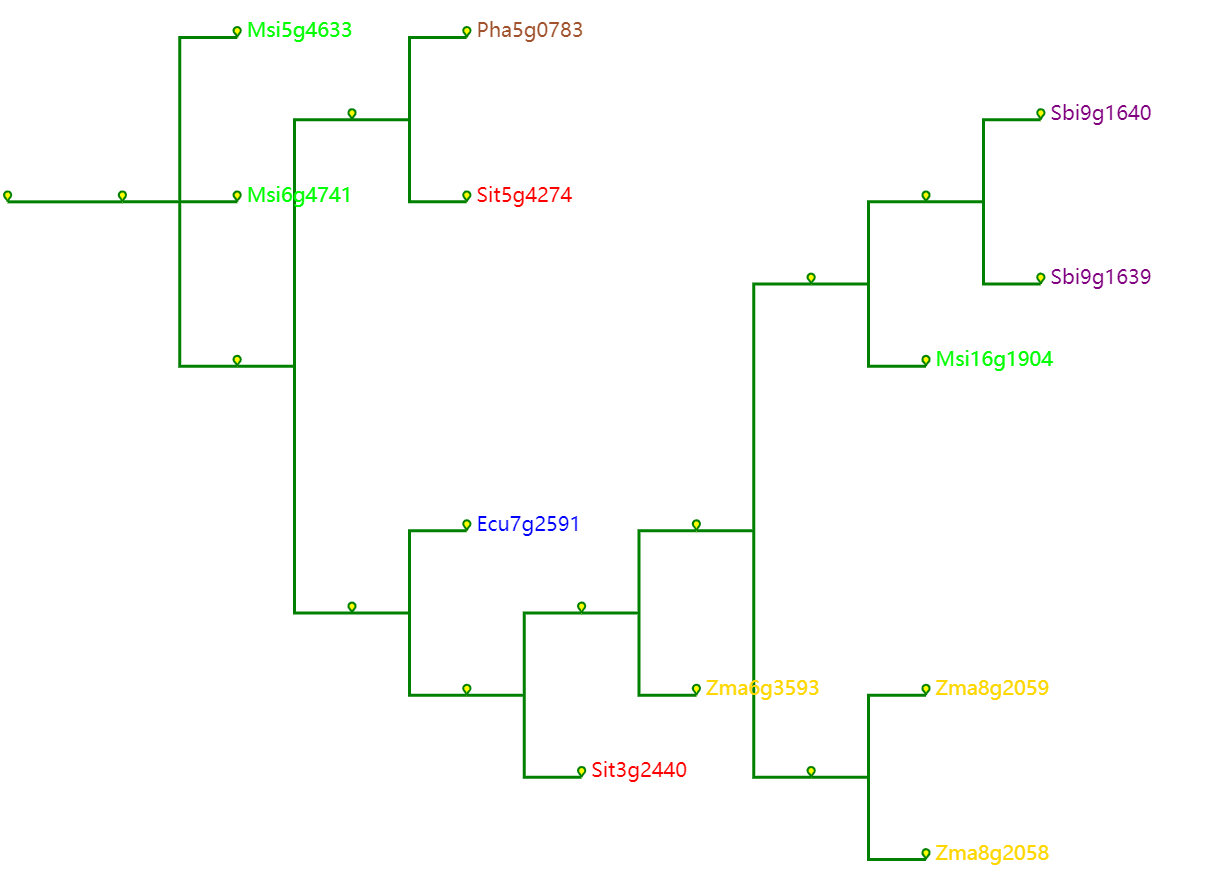
-
Ideal gene tree